This week the group finished working on their personal data collection, paper, and paper review of the other group. Because I ran my qPCR late last week, most of my effort was put towards working on the group paper last weekend and reviewing the salmon group's paper. In addition, after another group member ran the housekeeping gene as a reference I was able to calculate my Fold/Min Normalized Gene Expression Value and the data is below:
| Excel Data to calculate fold/min normalized gene expression value |
| Average gene expression in fold per min of HSP70 in control, V. tubiashii, copper, and V. tubiashii + copper treatments. |
NOTE: some values in column AC and AD were a little off, they have been corrected on the Google Sheet -
After viewing the comments on our paper, the remainder of my time will be to finish up and revise my sections of the paper as well as work on my ppt slides for our presentations on Wednesday.
It's been a fun quarter and I've learned a lot! Thanks for a great quarter and all the help with our projects!
Tuesday, November 30th, 2010:
RNA was extracted and quantified from half of the C. gigas gill tissue/per animal (the other half was used for Paul's protein extraction) using the following protocol:
RNA Extraction
- Turn on heating block and heat to 55°C.
- Incubate previously isolated RNA tissue sample tube at room temperature for 5 minutes.
- In the fume hood, add 200 uL of chloroform to sample making sure to close the tube before removing from fume hood.
- Vortex sample for 30 seconds so the sample solution becomes a milky emulsion.
- Incubate sample at room temperature for 5 minutes.
- Spin tube in refrigerated microfuge for 15 minuets at max speed.
- Gently remove tube from microfuge making sure to not to disturb the tube.
- Transfer most of the aqueous phase (top, clear portion) to a fresh microfuge tube labeled "Sam 10/12 RNA Ext C. gigas"
- Quickly close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself in proper chloroform disposal.
- Add 500uL isopropanol to the tube containing RNA and cap the tube.
- Invert the tube several times until the solution appears uniform (it should no longer appear lumpy).
- Incubate at room temperature for 10 minutes.
- Spin in refrigerated microfuge at max speed for 8 minutes making sure to place the tube in the microfuge so the tube is hinge pointing up, away from the center of the microfuge (this allows for optimal pellet formation).
- Remove supernatant making sure not to disturb RNA pellet.
- Add 1 mL of 75% EtOH to pellet.
- Close tube and vortex briefly to dislodge pellet from the side of the tube.
- Spin sample tube in refrigerated microfuge at 7500 g for 5 minutes.
- Carefully remove supernatant making sure again to not disturb pellet.
- Briefly spin tube for about 15 seconds.
- Pipette remaining EtOH.
- Leave tube open to allow pellet to dry at room temperature for no more than 5 minutes.
- Add 100 uL of 0.1% DEPC-H2O to sample tube and resuspend pellet by pipetting up and down.
- Incubated tube at 55°C for 5 minutes.
- Remove tube from heat, flick to mix, and place stock RNA sample on ice until quantified using Nanodrop spectrophotometer.
RNA Quantification
- To blank instrument, pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedestal and lower the arm and click blank on computer program.
- Wipe instrument with KimWipe, pipette 2 µL of RNA sample onto the Nanodrop pedestal and lower the arm, and click measure.
- Record your RNA concentration, A260/280 ratio, and A260/230 ratio.
- Raise the instrument's arm and wipe off sample with a KimWipe.
- Store RNA sample at -80°C.
RNA concentrations ~ 3000 ng/µL. Using (C1)(V1)=(C2)(V2), we normalized our samples by calculating how much concentrated RNA is needed to make a 500 µL of 200 ng/µL RNA stock from our initial concentrations.Because our samples were still very high in RNA concentration, we DNased 50 µL of all samples, individually, by following the Turbo Free DNase kit protocol in hopes to get rid of genomic DNA and other RNA contaminants left in our samples. Following this, the Nanodrop spectrometer was used to find the concentration of our samples again. Below are our results:
| Sample Number |
Control [RNA] (ng/µL) |
Cu [RNA] (ng/µL) |
Vt [RNA] (ng/µL) |
Cu+Vt [RNA] (ng/µL) |
| 1 A |
769.9 |
812.7 |
1479.1 |
1226.0 |
| 2 A |
1355.2 |
1304.7 |
1297.8 |
1403.1 |
| 3 A |
930.7 |
1319.8 |
1649.6 |
1382.1 |
| 4 A |
817.0 |
1764.3 |
1387.0 |
(no sample) |
| 1 B |
1119.8 |
940.8 |
863.1 |
885.5 |
| 2 B |
594.2 |
1567.1 |
1454.5 |
1052.8 |
| 3 B |
1514.8 |
1941.4 |
2214.9 |
1529.2 |
| 4 B |
(no sample) |
1524.1 |
1025.8 (not much sample is left but there's plenty of RNA in what we have) |
1030.1 |
With our 260/280 falling between 1.88-2.02, most around 1.91.
A final normalization step was taken for a final end concentration of 2 ug using the following equations:
1. (RNA conc ng/uL)/1000= RNA conc ug/uL
2. (RNA conc ug/uL) * X = 2 ug ---> X = 2 ug/(RNA conc ug/uL) where X equals the amount of stock RNA to add to the reverse transcription for RNA
The following google doc is the data and calculations for each sample: https://spreadsheets.google.com/ccc?key=t7fhoi_PCVo_3vcUnIIPQAA&hl=en#gid=0
*Note because final RNA amounts vary for each sample the amount of nuclease free H2O will vary as well and was calculated by water=(5-RNA)+4 and these calculations are included in the above spreadsheet as well*
All samples were reverse transcribed using the following protocol:
- Thaw and mix RNA stock tube by inverting
- Add pre-calculated stock RNA from google doc, 1 μl oligo dT, and individual amount of nuclease free H20 based off of RNA amount added in a 0.5 ml PCR tubes (also in google doc)
- Return RNA stock sample to ice
- Incubate mixture for 5 min at 70°C on the thermocycler then put tube on ice till cool.
- Briefly centrifuge sample tube
- Add 5 μl of M-MLV 5X Reaction Buffer, 5 ul of dNTPs, 1 μl of M-MLV RT, 4 μl of nuclease free H2O to sample tube.
- Incubate mixture for 60 minutes at 42°C and then heat inactivate at 70°C for 3 minutes on the thermocycler.
After reverse transcription each sample was transferred to its own individual labeled 1.5 mL microcentrifuge tube and an additional 225 uL of nuclease free water was added for a final dilution so enough product is available for qPCR's of three different genes in duplicates
All samples were returned to the -20°C for storage.
On Thursday, I set up my qPCR to examine HSP70 gene expression in C. gigas in the presence and absence of Vt and Cu using our previously transcribed cDNA samples using the following ingredients:
For a 25μl reaction volume:
| Component |
Volume |
Final Conc. |
| Master Mix, 2X (Immomix) |
12.5µL |
1x |
| Syto-13 dye (50uM) |
1µL |
2µM |
| HSP 70 forward primer, 10μM |
1.25μl |
2.5μM |
| HSP 70 reverse primer, 10μM |
1.25μl |
2.5μM |
| Ultra Pure Water |
7uL |
NA |
2 uL cDNA was used as a template, while 2 uL pure H20 was used as a negative control in the following plate map:
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
|
| A |
Control 1A |
Control 2A |
Control 3A |
Control 4A |
Control 1B |
Control 2B |
Control 3B |
NO SAMPLE |
| B |
Vt 1A |
Vt 2A |
Vt 3A |
Vt 4A |
Vt 1B |
Vt 2B |
Vt 3B |
Vt 4B |
| C |
Cu + Vt 1A |
Cu + Vt 2A |
Cu + Vt 3A |
NO SAMPLE |
Cu + Vt 1B |
Cu + Vt 2B |
Cu + Vt 3B |
Cu + Vt 4B |
| D |
Cu 1A |
Cu 2A |
Cu 3A |
Cu 4A |
Cu 1B |
Cu 2B |
Cu 3B |
Cu 4B |
| E |
Negative Control 1 |
Negative Control 2 |
Negative Control 3 |
Negative Control 4 |
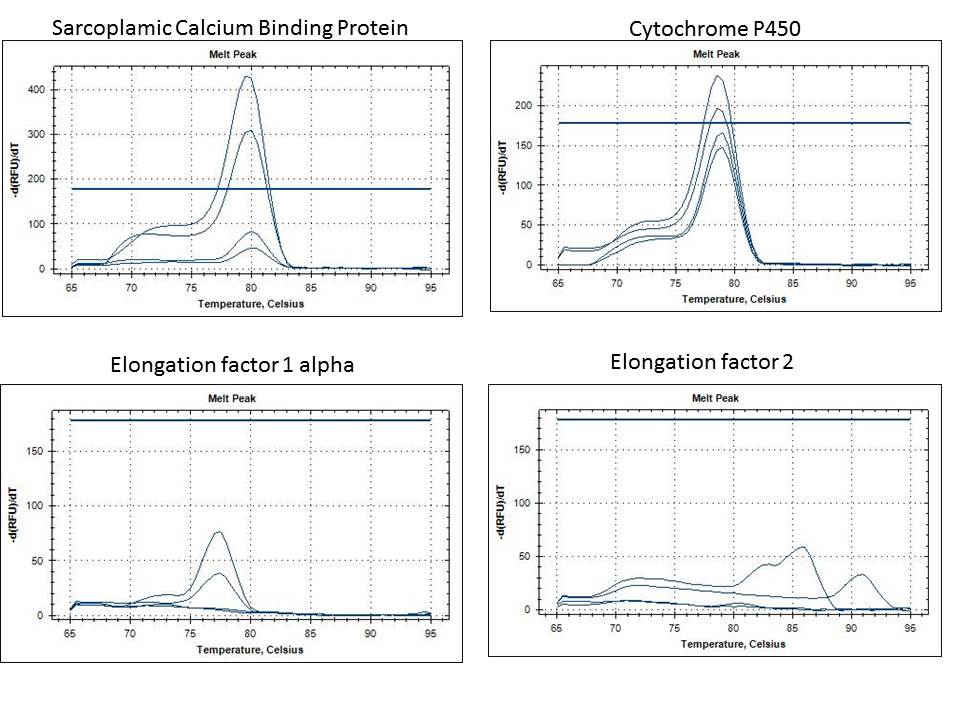
Results: I briefly looked at my qPCR results and there looks to be amplification in all samples except the negative control. Two samples (one from Cu and on from Cu+Vt) have a low melting point. I am waiting on another group member to run a plate with the house keeping gene as a reference to analyze the data from my run.
Lastly, the experiment set-up was bleached and broken down early in the week.
Tuesday, November 23rd, 2010:
Snow week!! Most of us were out for the week do to the weather. I believe Dave was able to work on processing some of his Vt challenge data but all other samples remained in storage until we are able to process them after the holiday. Happy Turkey Day!
Tuesday, November 16th, 2010:
Lab time was used to prep the experiment and make sure all supplies were available. Also Dave, David, and I started making up the LB for the Vt challenge.
This is week is the week animals are subjected to Cu, Vt, or Cu+Vt treatments. Oysters will be challenged for either 72 hrs in Cu or 48 hrs followed by an additional 24 hrs with Vt. The following is a visual outline of our plan (created by Dave):
 |
| Slide1.jpg |
LB was made for Vt dosing using the following protocol:
10 g NaCl
10 g Bactotryptone
5 g Yeast Extract
Mix on stir plate up to 800 ml with D/W
pH to 7.0
Add D/W to 1 L
Autoclave
130 ppm Cu was added to each Cu treatment on appropriate days (it was supposed to be 1.3 ppm because it is EPA relevant but I made a math error and the I/group didn't realize it until I had dosed the animals).
Because of slow Vt growth, Vt and Vp cultures were combined by Dave into one 500ml flasks of LB
Vt dosing and sampling were done on time.
Sampling prodcedure was as follows:
Tasks
Oyster Shucker - Ross for the 1st half followed by Jason
Gill tissue - me
Hemolymph - David
Mantle - Paul
Size measurements - Jason
Water filtering - Dave
Sampling over all went well. We did have two animals that were dead, but they had appeared to have been dead for some time and not due to our challenge. Some Cu treatment animals have a visibly green mantle. All Cu treatment animals had very green shells.
Tuesday, November 9th, 2010
This week we formed our groups for our class project. I am in the oyster group and I will be working on examining HSP 70 gene expression in C. gigas in the presence of Cu, Vt, and Cu+Vt. Ross, Dave, and I set up the experimental system in the basement Wednesday after class.
November 11-12th: We collected ~ 40 animals from Big Beef Creek and Belfair State Park for our experiment between 3 and 4 pm. Animals were kept in Steven's truck in coolers and added to the experimental tanks Friday morning.
Proposal
Introduction
Oysters are a very economically and ecologically important species in Washington state. Non-native C. gigas farmed animals are the number one shellfish resource for the state and produced over $57, 750, 000 in 2000 alone. Previous studies have supported that increased CO2 levels will most likely have detrimental effects on calcifying marine organisms, particularly at the larval stage (Talmage 2009). Increased quantities of CO2 will most likely affect metamorphosis and rearing abilities of calcifying organisms. Specifically, C. gigas has shown slower development rates in the presence of higher CO2 levels (Talmage 2009). When reared under high CO2 conditions (pH 7.4), more than 70% of C. gigas larvae developed without a shell or only had partial shell (Kurihara 2008). With large scale global climate changes threatening the future health of our oceans, an experiment studying the negative effects ocean acidification can have an important species is a very powerful tool to inform the public.
Material and Methods
CollectionC. gigas animals could be collected from numerous different sites around the Puget Sound region (n=30/site) that have demonstrated different pH levels. Hood Canal, for instance, has demonstrated recent pH levels as low as 7.6 and could be used as our most acidic field site. More ambient sites could be used as a natural control.
Laboratory C02 Exposure
Animals in two treatments/site (n=5/bucket resulting in 2 replicates/site/treatment) would be exposed to 700 ppm and 1000 ppm CO2 for a 2050 and 2100 predictions. Another control treatment would be held in replicates for each site (n=5/bucket). Water temperatures would be held consistent, but if we wanted a secondary stressor we could increase the number of treatments and include warmer water temperatures as that secondary stresor.
qPCR
Gill and mantle samples would be sampled and tested for gene expression in general stress response genes: HSP 70 and CytP450. RNA from samples would be isolated, normalized, and reverse transcribed into cDNA. cDNA would then be used as a template for a qPCR reaction.
Timeline
The first week would be dedicated to oyster collection. Week two would be the start of C02 exposure, which would last until the end of week 3 when sampling would occur. Week 4 would be for lab work and data analysis. Writing can be done at all stages.References
- Talmage, S.C. and Gobler, C.J. 2009. The effects of elevated carbon dioxide concentrations on the metamorphosis size, and survival of larval clams (Mercenaria mercenaria), bay scallops (Argopecten irradians), and Eastern oysters (Crassostrea virginica). Limol. Oceanogr. 54(6): 2072-2080.
- Kurihara, H. 2008. Effects of Co2-driven ocean acidification on the early developmental stages of invertebrates. Marine Ecology Progress Series. 373: 275-284.
Tuesday, November 2nd, 2010:Quantitative PCR and Epigenetics (continued)
Summary
- We will visualize our dot blot started in last weeks lab using westernbreeze chromogenic immunodetection. In addition, we will run a qPCR using the primers we have previously designed.
Materials and Methods
- Prepare 20 mL of Blocking Solution with the following:
- a. Ultra filtered Water 14 ml
- b. Blocker/Diluent (Part A) 4 ml
- c. Blocker/Diluent (Part B) 2 ml
- d. Total Volume 20 ml
2. Place membrane we prepared from last week in 10 ml of Blocking Solution in a plastic dish and cover
- 3. Incubate covered for 30 minutes on rotary shaker set at 1 revolution/second
- 4. Decant the Blocking Solution
- 5. Rinse membrane with 20 ml of water for 5 minutes. Decant water and repeat rinse.
- 6. Prepare 10 mL of Primary Antibody Solution (a 1:5000 dilution) with the following:
- a. Blocking Solution 10 ml
- b. 5-MeC antibody 2 µl
- c. Total Volume 10 ml
- 7. Incubate the membrane with 10 ml of Primary Antibody Solution for 1 hour on rotary shaker
8. Decant primary antibody. Then wash the membrane for 5 minutes with 20 ml of TBS-T.
10. Wash the membrane for 3 mins with 20 ml of TBS-T. Decant and repeat two more times
11. Rinse the membrane with 20 ml of water for 2 minutes and decant.
12. Incubate the membrane in 5 ml of Chromogenic Substrate until color begins to develop. (Color started to develop ~15 minutes after class)
Primer Reconstitution
- We needed to rehydrate our primers that arrived. In order to do so the nM measurement needs to have the decimal moved one decimal to the right and that amount of sterile water was added to each primer based on it's individual nM. This now is a 100 uM stock.
- We then created a 1:10 primer dilution by mixing 10ul primer and 90ul water in a new sterile tube.
qPCR
- Prepare master mix: Prepare enough master mix for SEVEN reactions to ensure sufficient volume recovery.
For a 50μl reaction volume:
| Component |
Volume |
Final Conc. |
- || Master Mix, 2X (Immomix) || 25µL || 1x ||
- || Syto-13 dye (50uM) || 2µL || 2µM ||
- || upstream primer, 10μM || 2.5μl || 2.5μM ||
- || downstream primer, 10μM || 2.5μl || 2.5μM ||
- || Ultra Pure Water || 16uL || NA ||
- 2. Add mastermix to each well of a white PCR plate (6 wells: 2 RNA, 2 cDNA, 2 negative)
- 3. Thaw cDNA samples and add 2uL cDNA template to appropriate wells.
- Add 2 uL RNA samples to appropriate wells.
- 5. Add 2uL of ultra pure water to the negative control wells.
6. Cap the wells securely.
7. Spin the strips
8. Ensure the lids are clean and place strips on ice
9. Our samples will be loaded in the qPCR machine later.
Results
- WesternBreeze® CHROMOGENIC IMMUNODETECTION
- qPCR
- (i'm having problems inserting this image. wiki doesn't want to upload my jpg file even though it's in the right format :-( )
Conclusions
- Dot Blot:
- The dot blot worked well for my grey whale DNA samples. There appears to be an appropriate color reduction as the dilution amounts increase.
- qPCR:
- Well you can't see it in my notebook but my qPCR looked great! My samples had a little genomic DNA contamination as shown by the melting temp graph and amplification graph, but the cDNA samples were stacked on top of each other perfectly!
- Reflection
- The dot blot worked well. It was interesting because I have never run that procedure prior to this lab so it was nice for the experience. The qPCR was my favorite part because I will be working on qPCR's soon in the Friedman Lab so this gave me some more practice and other than the genomic DNA contamination (that can be removed using DNase) it was awesome for my first solo qPCR.
Tuesday, October 26th, 2010: Epigenetics
Summary
- We will visualize our PCR reaction product (cDNA) from last weeks lab. In addition, we will preform a cytosine methylation dot blot in order to view DNA samples using westernbreeze chromogenic immunodetection.
Materials and Methods
Gel Electrophoresis
- Place previously made gel in gel box and cover with 1x TAE buffer
- Remove combs from wells
- Load 7uL 100 bp ladder in far left lane for visual aid

- Load 25 uL of PCR sample into each well and store remainder of product at -20ºC
- Run gel at at 100V for 55min, 150V for 10min, and 85V for 20min
- Visualize the gel on the UV transilluminator
DNA Dilution
- Prepare the following dilutions of your DNA (I signed up for the grey whale DNA)
| Dilution |
TARGET
|
ul of H20 |
ul of 20X SSC |
ul of 50ng/ul
|
- || 1 || 0.8 ng/ul || 124 || 60 || 16 ||
- || 2 || 0.4 ng/ul || 132 || 60 || 8 ||
- || 3 || 0.2 ng/ul || 136 || 60 || 4 ||
- || 4 || 0.1 ng/ul || 138 || 60 || 2 ||
- || 5 || 0.05 ng/ul || 139 || 60 || 1 ||
RNA Quantification
- To blank instrument, pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedestal and lower the arm and click blank on computer program.
- Wipe instrument with KimWipe, pipette 2 µL of RNA sample onto the Nanodrop pedestal and lower the arm, and click measure.
- Record your RNA concentration (=736.6 ng/µL), A260/280 ratio (=1.94), and A260/230 ratio (=1.57).
- Raise the instrument's arm and wipe off sample with a KimWipe.
- Store RNA sample at -80°C.
Dot Blotting
- Cut nylon membrane to fit the 72 wells of manifold
- Cover and soak nylon membrane in 6X SSC for 10 min
- Cut filter paper to the same size of the nylon membrane and wet using 6X SSC
- Assemble manifold with the membrane on top of filter paper
- Boil DNA in water for 10 minutes to denature and then transfer to ice to cool
- Turn on vacuum.
- Pipette 500 ul 6X SSC into each well and filter through.
- Centrifuge DNA for 5 minutes
- Map out samples
- Transfer all DNA to individual wells making sure not to touch membrane
- Filter DNA completely
- Soak cut filter in denaturation buffer
- Once filtration is complete, dismantle manifold and transfer membrane to filter paper soaked in denaturation buffer
- Let sit for 5 minutes
- Dry membrane on dry filter paper
- Wrap dried membrane in plastic wrap
- With DNA-side-down, view membrane on UV transluminator for 2 minutes at 120kJ to immobilize DNA
Results
- PCR:
 |
| PCR Gel Map |
-
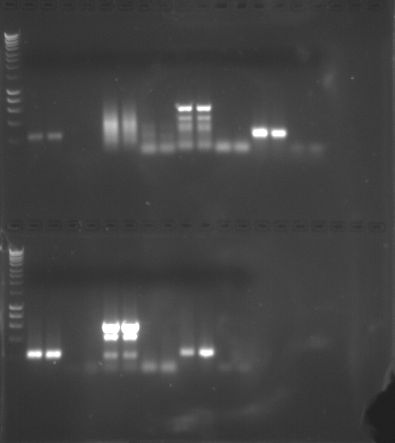
PCR Gel 1 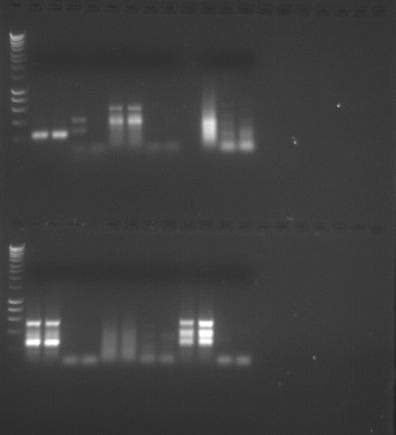
PCR Gel 2
Conclusions
- PCR:
- There appears to be some noise in my samples, possibly due to primers having a lower specificity, but there does seem to be two distinct bands at ~400 bp.
- There is some primer dimer in my samples creating a haze (see above about specificity).
- Negative controls were negative so contamination absent.
- Reflection
- Running the gel using our PCR cDNA product was useful. I have run them before but practice makes perfect and I enjoy seeing the results of the PCR. The dot blot procedure went well and it was nice that everyone got to take a step in the process for hands on experience. Next week we will finish up the procedure by completing the westernbreeze chromogenic immunodetection.
Tuesday, October 19th, 2010: Reverse transcription and end-point PCR
- Using our RNA stock samples we extracted last week, RNA was reverse transcribed into cDNA which was then used to amplify a gene of interest usinf end-point PCR. In addition, we prepared our agrose gels for next weeks lab where we will use it to visualize our PCR results.
- Materials and Methods
Reverse Transcription
- Thaw and mix RNA stock tube by inverting
- Add 5 μl stock RNA, 1 μl oligo dT, 4 μl of nuclease free H20 in a 0.5 ml PCR tube labeled with your "Sam cDNA”
- Return RNA stock sample to ice
- Incubate mixture for 5 min at 70°C on the thermocycler then put tube on ice till cool.
- Briefly centrifuge sample tube
- Add 5 μl of M-MLV 5X Reaction Buffer, 5 ul of dNTPs, 1 μl of M-MLV RT, 4 μl of nuclease free H2O to sample tube.
- Incubate mixture for 60 minutes at 42°C and then heat inactivate at 70°C for 3 minutes on the thermocycler.
- Spin down the sample in a desk top centrifuge and store on ice or at -20°C.
Polymerase Chain Reaction
- In a tube labeled "SB MM" add 250 μl GoTaq®Green Master Mix 2X, 15 μl of cytochrome oxidase P450 foward primer, 15 μl of cytochrome oxidase P450 reverse primer, 108 μl of nuclease free H2O to make master mix.
- Label 4 0.5 mL PCR tubes "(1-4) SB"
- Pipette 48 μl master mix in to each of the 0.5 mL PCR tubes
- Add 2 μl of the corresponding DNA template to each tube and mix by pipetting up and down.
- cDNA
- cDNA
- nuclease free H2O
- nuclease free H2O spectrophotometer.
- Cap all tubes tightly, spin to pool liquid, and load reactions into thermocycler
- Put samples through the following thermal cycling profile.
| Step |
Temperature |
Time |
Cycles |
- || Denaturation || || 95C|| || 5 min || || 1||
- || Denaturation || || 95C|| || 30 sec || || 40 ||
- || Annealing || || 55C|| || 30 sec ||^ ||
- || Extension || || 72C|| || 90 sec ||
- || Final extension || || 72C|| || 3 min || || 1||
- || Hold || || 4C|| || ∞|| ||= 1 ||
- Stored at -20°C.
- Mix 2g of agarose with 150mL 1x TAE in a 1L flask
- Microwave solution for ~ 3 minutes swirling occasionally
- Cool solutionand add 12 uL ethidium bromide (EtBr) and mix by swirling
- Pour into gel tray, add gel combs, and allow to set and cool
- After gel is set, wrap in plastic and foil wrap and place in fridge for next week's lab.
Results
- Our agrose gels set nicely and were stored for next weeks lab to visualize our PCR results using gel electrophoresis.
Conclusions
- No conclusions can be drawn yet until we run our PCR samples through agrose gel electrophoresis.
- Reflection
- This lab helped re-familiarize me with making gels and preparing our samples for gel electrophoresis. Prior to this lab, I had never completed a reverse transcription so I'm excited to see the amplification visual results of on the agrose gel next week when we run the gel electrophoresis. I also didn't know you could pre-make your agrose gels and store them in the freezer for future use.
Tuesday, October 12th, 2010: RNA Extraction and Protein Analysis, Part 2
- Using the same two different samples of C. gigas gill tissue from last weeks lab, RNA was extracted and quantified from the tissue sample we isolated from last week, while the protein we extracted from the other gill tissue was separated using an SDS-PAGE gel.
Materials and Methods
SDS-Page Protocol
- Thaw previously frozen protein stock sample using body heat by rolling tube in hands for 1-2 mins
- Add 15uL of protein stock and 15uL of 2X Reducing Sample Buffer to a new 1.5mL screw cap tube labeled "Sam 10/12 protein + buffer".
- Return protein stock to -20°C freezer
- Flick protein + buffer tube to mix.
- Briefly centrifuge for ~10 seconds.
- Boil sample for 5 minuets in water boiling water bath.
- Assemble of gel box and gels while boiling making sure to rinse gel wells thoroughly.
- After boiling, centrifuge for 1 minute.
- Load entire sample into the appropriate well using appropriate gel loading tip. Deviation from protocol: some bubbles forced a small amount of protein + buffer sample out of well.
- Put lid on gel box and plug electrodes into appropriate receptacles on the power supply.
- Turn power supply on and set voltage to 150V. Run gel for 45 minutes.
- Turn off power supply and disconnect gel box from power supply, remove lid from gel box, disengage the tension wedge, remove gel from gel box.
- Carefully crack open cassette to expose gel and trim wells at top of gel. Make sure to notch a corner to remember correct orientation of gel.
- Place gel in a designated container and cover the gel with Coomassie Stain.
- Incubate gently on shaker for 5 minutes.
- Return Coomassie Stain to original container.
- Rinse gel briefly with 10% acetic acid and pour this wash down the drain.
- Continue to replace and add 10% acetic acid to container with gel while incubating on shaker every 15 minutes until bands become clearly visible. (This may need to incubate overnight. If so, cover container with plastic wrap and leave on shaker)
- Once bands are visible take picture of gel.
RNA Extraction
- Turn on heating block and heat to 55°C.
- Incubate previously isolated RNA tissue sample tube at room temperature for 5 minutes.
- In the fume hood, add 200 uL of chloroform to sample making sure to close the tube before removing from fume hood.
- Vortex sample for 30 seconds so the sample solution becomes a milky emulsion.
- Incubate sample at room temperature for 5 minutes.
- Spin tube in refrigerated microfuge for 15 minuets at max speed.
- Gently remove tube from microfuge making sure to not to disturb the tube.
- Transfer most of the aqueous phase (top, clear portion) to a fresh microfuge tube labeled "Sam 10/12 RNA Ext C. gigas"
- Quickly close the tube containing the organic and interphase and properly dispose of the liquid inside the tube as well as the tube itself in proper chloroform disposal.
- Add 500uL isopropanol to the tube containing RNA and cap the tube.
- Invert the tube several times until the solution appears uniform (it should no longer appear lumpy).
- Incubate at room temperature for 10 minutes.
- Spin in refrigerated microfuge at max speed for 8 minutes making sure to place the tube in the microfuge so the tube is hinge pointing up, away from the center of the microfuge (this allows for optimal pellet formation).
- Remove supernatant making sure not to disturb RNA pellet.
- Add 1 mL of 75% EtOH to pellet.
- Close tube and vortex briefly to dislodge pellet from the side of the tube.
- Spin sample tube in refrigerated microfuge at 7500 g for 5 minutes.
- Carefully remove supernatant making sure again to not disturb pellet.
- Briefly spin tube for about 15 seconds.
- Pipette remaining EtOH.
- Leave tube open to allow pellet to dry at room temperature for no more than 5 minutes.
- Add 100 uL of 0.1% DEPC-H2O to sample tube and resuspend pellet by pipetting up and down.
- Incubated tube at 55°C for 5 minutes.
- Remove tube from heat, flick to mix, and place stock RNA sample on ice until quantified using Nanodrop spectrophotometer.
RNA Quantification
- To blank instrument, pipette 2µL of 0.1%DEPC-H20 onto the Nanodrop pedestal and lower the arm and click blank on computer program.
- Wipe instrument with KimWipe, pipette 2 µL of RNA sample onto the Nanodrop pedestal and lower the arm, and click measure.
- Record your RNA concentration (=736.6 ng/µL), A260/280 ratio (=1.94), and A260/230 ratio (=1.57).
- Raise the instrument's arm and wipe off sample with a KimWipe.
- Store RNA sample at -80°C.
Protein Gel Key:
Protein Gel 1:
| Protein Gel 1 |
Protein Gel 2:
| Protein Gel 2 |
- RNA extraction and quantification from 37 mg C. gigas gill tissue sample:
- RNA concentration = 739.6 ng/µL
- A260/A280 = 1.94
- A260/230 = 1.57
Conclusions
- SDS-PAGE gel:
- All the protein samples loaded in both gels appear to have run well. All shared species have similar band patterns, except for well 9 on gel 1 and well 2 on gel 2. Both are pH treated sockeye salmon, but well 9 has a major band that does not appear in the other sockeye samples, while well 2 on the other gel appears to have run off the gel. Both could have been due to over loading the well initially while loading the protein sample. Good. -
storercg
- All the protein samples loaded in both gels appear to have run well. All shared species have similar band patterns, except for well 9 on gel 1 and well 2 on gel 2. Both are pH treated sockeye salmon, but well 9 has a major band that does not appear in the other sockeye samples, while well 2 on the other gel appears to have run off the gel. Both could have been due to over loading the well initially while loading the protein sample. Good. -
- RNA extraction and quantification:
- My extraction falls within the acceptable range of 1.8-2.0 for the A260/A280 ration and also falls withing the A260/A230 range of 1.5-2.0, which implies my RNA is pure and clean.
- SDS-PAGE gel:
- Reflection
- The purpose of this lab was to familiarize ourselves with RNA extraction and quantification as well as an introduction to visualizing our extractions using a protein SDS-PAGE gel. Quantifying the RNA also allowed us to test its purity. In the future, we could amplify the RNA or run a qPCR. The neat tool I learned about in this lab was the pre-made gels. I never knew these existed prior to this lab. I also learned that bubbles can force your protein sample out of its well if you are not careful.
Tuesday, October 5th, 2010: RNA Extraction and Protein Analysis, Part 1
- Using two different samples of C. gigas gill tissue, RNA was isolated from the first sample with the additon of TriReagent, while protein was extracted from the other gill tissue sample with the addition of CellLytic MT. The protien extracted from the second sample wsa then quantified to determine the concentration of protiens in the sample using the Bradford Assay.
Materials and Methods
RNA Isolation
- Microcentrifuge tube with tissue sample labeled "C. gigas gill tissue B 37 mg Sam 10/5 " (note: the sample was stored on ice when not working directly on the homogenizing the tissue)
- 500 µL of TriReagent added to tube with tissue sample
- Gill tissue was homogenized using a sterile pestle. Brief vortexing was used in order to ensure homogenization.
- An additional 500 µL of TriReagent was added to homogenized tissue.
- Sample was vortexed for 15s and then stored in -80°C until next week where I will finish the RNA extraction.
Protein Extraction
- Microcentrifuge tube with tissue sample labeled "C. gigas gill tissue B 22 mg Sam 10/5 " (note: the sample was stored on ice when not working directly on exctracting protien from the tissue)
- 500 µL of CellLytic MT added to tube with tissue sample
- Gill tissue was homogenized using a sterile pestle. Deviation from protocol: Brief vortexing was used to homogenize the sample. Nice note! -
storercg
- Tube was inverted several times
- Tissue sample was spun down in a refrigerated microfuge for 10 minuets at max speed.
- Approximatley 45 µL of supernatent was pipetted off from the tube and transfered into a new tube labeled "Protien, C. gigas gill B, Sam, 10/5"
- Both tubes were stored on ice until moved into the -20°C freezer
Protein Quantification using Bradford Assay
- New 2 mL tube labeled "Protien BA Sam 10/5 "
- 15 µL of protein supernatant from tube labeled "Protien, C. gigas gill B, Sam, 10/5" and 15 µL of DI water was pipetted into the new 2 mL tube to dilute the original protein sample 1:2
- A blank sample was created by pipetting 30 µL of DI water into another 2 mL tube labeled "Blank, Sam, 10/5"
- Both 2 mL tubes were mixed using pipetting
- 1.5 mL Bradford reagent was added to both tubes
- Both tubes were inverted several times for mixing purposes and allowed to incubate at room temperature for 10 minutes, inverting both tubes several times during incubation
- Both tubes were mixed via pipetting and then transferred to their individual plastic cuvette
- Using a spectrometer, the blank sample was used to zero the machine
- The cuvette containing the 1:2 protein dilution was then placed in the spectrometer and the absorbance value was measured twice at 595nm, pipetting the sample in between measurements (measurement1 =0.173, measurement2 =0.170)
- The two absorbance values were averaged using the following equation
(0.173+0.170)/2=0.171511.The protein concentration (y) was then calculated using the following equation from the Pierce Coomassie (Bradford) Protein Assay Kit (shown below equation): y=[1019.9(0.1715) X 2]
y=347.8 ug/mL
 1212. All protein sample tubes (supernatant and original homogenized sample) were returned to TA and stored in the -20°C freezer
1212. All protein sample tubes (supernatant and original homogenized sample) were returned to TA and stored in the -20°C freezer
Primer Design
- Great job mining the EST database!! Most of the genes you have posted are putative and since there are annotated (identified) HSP70 and glutathione peroxidase genes for the Pacific Oyster available I recommend using those sequences instead. For a more detailed explanation please see each gene. -
storercg
- Below are three sequences for three different genes for C. gigas found from NCBI. It looks like
- Cellular respiration -- NADH dehydrogenase subunit 4 Since all of these are mRNA sequences they should be fine for primer design. These were genes identfied to either up regulated or down regualated so they might be quite interesting. However, the gene identity is putative, indicated by "similar to" so I would not recommend using this gene or putative sequences for primer design in this class (see explanation above). -
storercg
- EST638 subtracted libraries from oysters exposed to temperature challenge Crassostrea gigas cDNA similar to NADH dehydrogenase (ubiquinone) Fe-S protein, mRNA sequence
243 bp linear mRNA
EE677777.1 GI:164111806ESTGenBankFASTARemove from clipboard2.EST117 subtracted libraries from triploid versus diploid in autumn Crassostrea gigas cDNA clone U-3 similar to putative NADH dehydrogenase subunit 4L, mRNA sequence
292 bp linear mRNA
CX739454.1 GI:84142220ESTGenBankFASTARemove from clipboard3.EST37 subtracted libraries from oysters exposed to temperature challenge Crassostrea gigas cDNA similar to NADH dehydrogenase subunit 4, mRNA sequence
585 bp linear mRNA
DW713851.1 GI:150037218ESTGenBankFASTARemove from clipboard
- EST638 subtracted libraries from oysters exposed to temperature challenge Crassostrea gigas cDNA similar to NADH dehydrogenase (ubiquinone) Fe-S protein, mRNA sequence
- Stress response -- heat shock protein 70 Again any of these sequences will can be used for gene expression primer design since they are mRNA/cDNA, but they are putative. You might want to try searching "Pacific oyster HSP70". When I did this any of the top three hits (e.g. AF144646.1) would work and there is more confidence in the identity of these genes. -
storercg
- Ef1_4_31.G04_070123198Y Eisenia fetida cDNAs from SSH library Eisenia fetida cDNA similar to heat shock protein 70 [Crassostrea gigas] (AAD31042), Expect=6e-31, mRNA sequence
248 bp linear mRNA
EY892557.1 GI:209528987ESTGenBankFASTARemove from clipboard2.AUBA-aaa15d10.g2 A.suum_EST_64.5_Embryo Ascaris suum cDNA similar to gb|AAD31042.1|AF144646_1 heat shock protein 70 [Crassostrea gigas], mRNA sequence
338 bp linear mRNA
FE918139.1 GI:171289160ESTGenBankFASTARemove from clipboard3.BGBV-aae36a09.g1 Snail_EST_pSMART Biomphalaria glabrata cDNA similar to gb|AAD31042.1|AF144646_1 heat shock protein 70 [Crassostrea gigas], mRNA sequence
592 bp linear mRNA
EX006714.1 GI:156763330ESTGenBankFASTARemove from clipboard
- Ef1_4_31.G04_070123198Y Eisenia fetida cDNAs from SSH library Eisenia fetida cDNA similar to heat shock protein 70 [Crassostrea gigas] (AAD31042), Expect=6e-31, mRNA sequence
- Oxidative metabolism and the immune system -- glutathione peroxidase The same applies here as it did for HSP70. Check out EF692639.1. -
storercg
- EST80 subtracted libraries from oysters exposed to temperature challenge Crassostrea gigas cDNA similar to glutathione peroxidase, mRNA sequence
308 bp linear mRNA
DW713894.1 GI:150037131ESTGenBankFASTAItem in clipboard
- EST80 subtracted libraries from oysters exposed to temperature challenge Crassostrea gigas cDNA similar to glutathione peroxidase, mRNA sequence
- Cellular respiration -- NADH dehydrogenase subunit 4 Since all of these are mRNA sequences they should be fine for primer design. These were genes identfied to either up regulated or down regualated so they might be quite interesting. However, the gene identity is putative, indicated by "similar to" so I would not recommend using this gene or putative sequences for primer design in this class (see explanation above). -
- Below are three sequences for three different genes for C. gigas found from NCBI. It looks like
3.EST128 subtracted libraries from triploid versus diploid in autumn Crassostrea gigas cDNA clone V-8 similar to putative glutathione peroxidase, mRNA sequence
Results
- RNA isolation sample was stored for use in next weeks lab. 347.8 ug/mL of protein was extracted from a 22 mg piece of C. gigas gill tissue.
Conclusions
- I have no previous knowledge of protein quantification to compare my results to, but based off of the calculated protein concentration it appears that there is a sufficient amount of protein to continue with. Based on this, the next step we will take is to run a SDS-PAGE in hopes to separate proteins from one another based on their molecular weight. In addition, tree cDNA or mRNA sequences were found for three genes (NADH dehydrogenase subunit 4, heat shock protein 70, and glutathione peroxidase) for future instruction on primer design.
- Reflection
- The purpose of this lab was to familiarize ourselves with RNA and protein extraction as well as protein quantification. These tools will be very useful for us in our upcoming research project as well as in future research outside of this class. In addition NCBI was introduced to us as a resource for primer design. I am still working on familiarizing myself with this resource but was able to find the sequences requested for each gene of interest to me.
